| ID |
Date |
Author |
Type |
Category |
Subject |
|
2
|
Tue Feb 14 08:44:37 2023 |
Rutuja | Summary | Wiki | Setup logs for Chimay jupyterhub + web server setup |
This are my notes from Winter 2022 before the Richardson Lab ELOG was setup. TODO: Need to write a proper "HowTo" ELOG for jupyterhub + web server setup based on this.
JupyterHub on Chimay
[Early Jan 2022]
Cannot install using TLJH since it's only available for Ubuntu.
So instead, installing step-by-step using guide available here - https://github.com/jupyterhub/jupyterhub-the-hard-way/blob/master/docs/installation-guide-hard.md
Nginx webserver is used for HTTPS reverse-proxy for the jupyterhub.
I installed nginx via apt-get, config at - /etc/nginx/sites-available/default
I used cerbot to generate SSL certificates.
To start the webserver
sudo systemctl start nginx.service
Goal: Setup JupyterHub as a systemd service and make it available over HTTPS.
To start the services needed for the hub
sudo systemctl start jupyterhub.service
Note: Reload the daemon if you edited a service before you restart it, otherwise it will complain.
sudo systemctl daemon-reload
Logs
[Jan 26, 2020]
Ended up using the JupyterHub guide available here - https://github.com/jupyterhub/jupyterhub-the-hard-way/blob/master/docs/installation-guide-hard.md and SudoSpawner guide available here - https://jupyterhub.readthedocs.io/en/stable/reference/config-sudo.html
I was struggling with this quite a bit but eventually I figured out that I was running into trouble because I was trying to run jupyterhub command as a password-less sudo user as I thought that's what sudospawner needed. That seems not to be the case.
- The systemd service
jupyter.service has to be run as User=root and the ExecStart=/opt/jupyterhub/bin/jupyterhub -f /opt/jupyterhub/etc/jupyterhub/jupyterhub_config.py and ensure that /usr/sbin is in PATH. (This necessary as we need to spawn single-user jupyter servers for our users on-demand, so the User that runs the ExecStart needs to have permissions to run single-user server init commands.)
- In
jupyterhub_config.py, the spawner is configed as sudospawner - c.JupyterHub.spawner_class=sudospawner.SudoSpawner. I installed sudospawner in our jupytherhub environment via pip.
- In
jupyterhub_config.py, the authenticator used is PAM which allows linux users on chimay to login with there username and password and opens their shell in their home directories.
[Jan 27, 2020]
[10:30]
Attempted MATLAB integration following this guide - https://github.com/mathworks/jupyter-matlab-proxy
It annoyingly broke nodejs which broke jupyterhub as the proxy failed without it. I have now restored nodejs and the jupyterhub is back up.
[12:00]
User controls now has admin access on the hub. To add more admins - edit /opt/jupyterhub/etc/jupyterhub/jupyterhub_config.py by adding the username in c.Authenticator.admin_users
[15:15]
Argh! User's cannot access user-created conda environments in notebooks besides the default one i.e. the one for which kernelspec is available in /usr/local/share/jupyter/kernels. Non-sudo users also can't access this path to put a kernelspec in it! Sudo users who can do this end up having their custom environments visible to everyone. Argh!
!FIX NEEDED!
[15:54]
After reading this (https://github.com/Anaconda-Platform/nb_conda_kernels/issues/185) I am putting
"CondaKernelSpecManager": {
"kernelspec_path": "--user"
}
in /opt/miniconda3/etc/jupyter/jupyter_config.json
WebServer down, fix yet to be tested.
[16:04]
Fix does not seem to be working.
[Tue Feb 1 17:02:46 2022]
ISSUE: User unable to use their custom conda environments in notebooks
STATUS: Fixed, refer to elog 'ligo-ds' no. 1
All future logs will be on the ELOG. |
|
3
|
Wed Aug 9 18:15:25 2023 |
Rutuja Gurav | Infrastructure | Scripts/Programs | User unable to make matplotlib interactive plots |
| It seems like users other than `controls` are unable to make matplotlib interactive plots in jupyter notebooks using widgets in their python environments.
Users get an error message - "Error displaying widget: model not found ".
I tested this out on `controls` and `rutuja`.
Looking for a fix.
[Update] Found this git issue discussing a possible fix - https://github.com/matplotlib/ipympl/issues/486 |
|
7
|
Sun Feb 25 14:54:55 2024 |
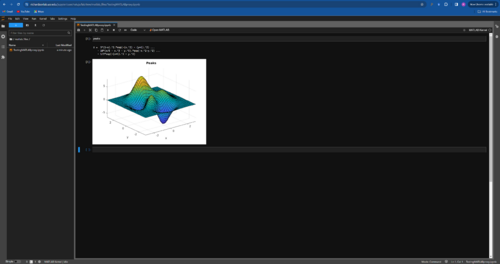
Rutuja Gurav | HowTo | Scripts/Programs | Using MATLAB on JupyterHub |
Prerequisites - Make sure you have your Mathworks login details handy. UCR provides a single-user MATLAB license to students.
Steps:
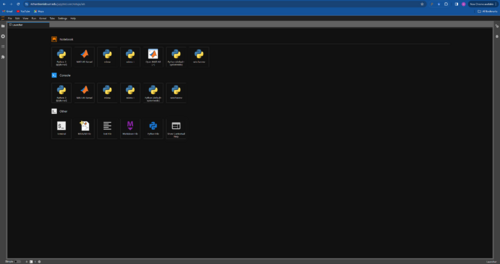
- Login to JupyterHub and select Open MATLAB from the Launcher tab. Click on Open MATLAB.
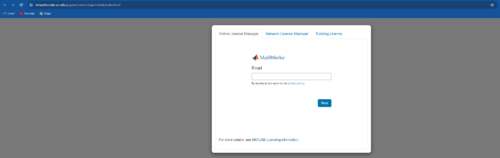
- Enter your Mathworks login detail in the Online License Manager window that pops up.
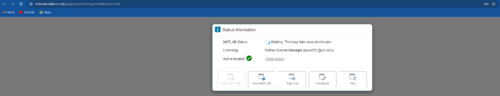
- Status Information window will popup and MATLAB startup process will begin. This can take a few minutes the first time. You'll eventually land on a standard MATLAB GUI.
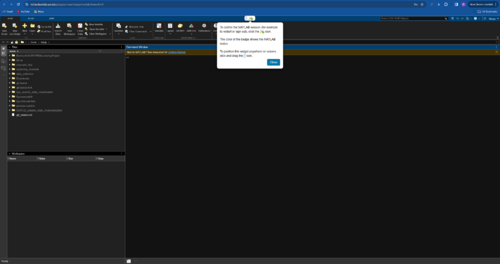
- From here on, you can continue developing in the MATLAB GUI or you can navigate back to your JupyterHub launcher to create a notebook and run MATLAB code in a notebook. Just choose MATLAB kernel to execute the code.
- You can close the MATLAB session and sign out by using the navigation button that pops up on the MATLAB GUI.
|
| Attachment 1: richardsonlab_jupyter_matlab_proxy_HowTo_0__user_rutuja.png
|
|
| Attachment 2: richardsonlab_jupyter_matlab_proxy_HowTo_1__user_rutuja.png
|
|
| Attachment 3: richardsonlab_jupyter_matlab_proxy_HowTo_2__user_rutuja.png
|
|
| Attachment 4: richardsonlab_jupyter_matlab_proxy_HowTo_3__user_rutuja.png
|
|
| Attachment 5: richardsonlab_jupyter_matlab_proxy_HowTo_4__user_rutuja.png
|
|
|
8
|
Tue Feb 27 18:00:01 2024 |
Pooyan | HowTo | Scripts/Programs | Install Matlab add-ons |
SIS requires two Matlab toolboxes (Parallel computing and Symbolic math), but a simple add-on installation by user fails due to not having write permission on " /usr/local/MATLAB/".
Theoretically, one can install the toolbox only on their own home directory; but changing the installation directory in settings didn't solve the problem. (Matlab still asking for write permission)
I tried giving user the appropriate access using the "setfacl". although all permissions are granted to the user, Matlab still complains about not having write permission.
Using a root access user works on a local machine, but fails in this situation due to X11 not being forwarded when run by super user. The add-ons can only be installed using the GUI, there is no easy CLI command for that.
Fix:
Used the fix on this blog [https://danct12.github.io/Fix-X11-Forwarding-sudo/] to add the current user's SSH session authentication to the root's .Xauth file.
$ echo $DISPLAY
$ xauth list $DISPLAY
$ sudo xauth add chimay/unix:12 MIT-MAGIC-COOKIE-1 c9acf72143c05334f58f**********220
after this, the user can run matlab as super user, "sudo matlab", and use the GUI. This should be done by a user that has sudo access.
This process seems to install the toolboxes for all users, so there shouldn't be any need of doing this by others.
|
|
10
|
Mon May 27 13:24:03 2024 |
Rutuja Gurav | Infrastructure | Scripts/Programs | Large data collection scripts just die without error |
| I can't quite figure out why my standard data collection scripts just die without any error logged in my console dump.
I just get the progress bar printed by NDS and it just stops in the middle of data download. The ETA is also unexpectedly high! I was able to download these very channels for O3b just fine but O3a download is being difficult. I could download the ISI-GND_STS BLRMS channels and the PEM WIND channels for both O3a and O3b just fine in reasonable time (<10 hrs).
Example data collection run console dump for ACC channels -
PROJECT_DIR: /home/rutuja/ligo_seismic_state_characterization
Config file loaded
Run config:
{'channels_list_path': 'data/channels_lists/L1/pem_acc_channels.txt',
'data_agg': 'rms',
'data_download_dir': 'data/download/L1',
'data_trend': 's-trend',
'end_time': '2019-08-01T00:00:00',
'gaps_pad': 'nan',
'ifo': 'L1',
'start_time': '2019-07-01T00:00:00'}
Period: 2019-07-01T00:00:00 to 2019-08-01T00:00:00
pem_acc_channels channels list loaded
61 channels to be used
Attempting to access data from frames...
unknown datafind configuration, cannot discover data
Failed to access data from frames, trying NDS...
Opening new connection to nds.ligo.caltech.edu... connected
[nds.ligo.caltech.edu] set ALLOW_DATA_ON_TAPE='True'
Checking channels list against NDS2 database... done
Querying for data availability... done
Found 1 viable segments of data with 100.00% coverage
Downloading data: | | 0/2678400.0 ( 0%) ETA ?
Downloading data: | | 0/2678400.0 ( 0%) ETA ?
Downloading data: |▌ | 137519.0/2678400.0 ( 5%) ETA 77:29:58
Downloading data: |█ | 275038.0/2678400.0 ( 10%) ETA 49:49:41
Downloading data: |█ | 275038.0/2678400.0 ( 10%) ETA 49:49:41
It just hangs here and the script is just killed, I think. I've been restarting this particular run for ages.
On other occasions, the script actually does end gracefully but that's because of a peculiar error and only partial data is downloaded!
PROJECT_DIR: /home/rutuja/ligo_seismic_state_characterization
Config file loaded
Run config:
{'channels_list_path': 'data/channels_lists/L1/pem_acc_channels.txt',
'data_agg': 'rms',
'data_download_dir': 'data/download/L1',
'data_trend': 's-trend',
'end_time': '2019-10-01T00:00:00',
'gaps_pad': 'nan',
'ifo': 'L1',
'start_time': '2019-09-01T00:00:00'}
Period: 2019-09-01T00:00:00 to 2019-10-01T00:00:00
pem_acc_channels channels list loaded
61 channels to be used
Opening new connection to nds.ligo.caltech.edu... connected
[nds.ligo.caltech.edu] set ALLOW_DATA_ON_TAPE='True'
Checking channels list against NDS2 database... done
Querying for data availability... done
Found 1 viable segments of data with 100.00% coverage
Downloading data: | | 0/2592000.0 ( 0%) ETA ?
Downloading data: | | 0/2592000.0 ( 0%) ETA ? read_server_response: Wrong length read (0)
Downloading data: | | 0/2592000.0 ( 0%) ETA ?
Elapsed time: 5:15:47.576076
Data spans 2019-09-01 00:00:00 to 2019-10-01 00:00:00
Saving data to: /home/rutuja/ligo_seismic_state_characterization/data/download/L1/pem_acc_channels/agg_rms--trend_s-trend--gaps_nan/2019-09-01T00:00:00--2019-10-01T00:00:00.hdf5
On chat.ligo.org, someone said there was nothing I could do about the read_server_response: Wrong length read (0) error... |
|
11
|
Sun Jun 2 15:53:47 2024 |
Rutuja Gurav | Infrastructure | Scripts/Programs | Unable to access Segments DB on Chimay? |
Cannot access SegDB on Chimay. See error below.
from gwpy.segments import DataQualityFlag
from gwpy.time import to_gps, from_gps
ifo, run = 'L1', 'O4'
if run == 'O3':
O3a_start, O3a_end = '2019-04-01T08:00:00', '2019-10-01T08:00:00'
O3b_start, O3b_end = '2019-11-01T08:00:00', '2020-03-27T10:00:00'
start_gps, end_gps = to_gps(O3a_start), to_gps(O3b_end)
if run == 'O4':
O4_start, O4_end = '2023-05-24T15:00:00', '2024-01-16T16:00:00'
start_gps, end_gps = to_gps(O4_start), to_gps(O4_end)
print(start_gps, end_gps)
segs = DataQualityFlag.query(ifo+':DMT-ANALYSIS_READY:1',start_gps, end_gps, url="https://segments.ligo.org")
HTTPError: 401 Client Error: Unauthorized for url: https://segments.ligo.org/dq/L1/DMT-ANALYSIS_READY/1?e=1389456018&include=metadata,known,active&s=1368975618
|
|
1
|
Tue Feb 1 17:02:46 2022 |
Rutuja | Update | ELOG | JupyterHub Setup: Users unable to use their conda environments in jupyter notebooks |
Issue: Currently, the only kernelspec present in /usr/local/share/jupyter/kernels is the miniconda environment called 'python' (/opt/miniconda3/envs/python/).
Goal: We want users to be able to create their own conda environs and use them in their jupyter notebooks. Currently they can only use their custom environs in terminal.
ToDo: Figure out a way to place user1's custom environment 'mytestenv' kernelspec in /home/user1/.local/share/jupyter/kernels/mytestenv
[UPDATE] Fixed this issue. Users should follow the following steps to have their custom conda environs available in their jupyter notebooks.
1. Say you have a conda env named mytestenv. Activate this env.
2. Install ipykernel in the conda env you want to add to Jupyter:
$ conda install ipykernel
3. Install the kernel
$ python -m ipykernel install --user --name mytestenv --display-name "My Test Env"
You can always change the --name (how this kernel appears in your jupyter directory) and --display-name (how it appears in the JupyterLab's 'Select Kernel' window) but it is recommended to match it with your conda environment name to avoid future confusion.
4. Check if you installed the kernel successfully:
$ jupyter kernelspec list
[UPDATE: JR] After completing step 4, you must restart your server for the newly installed environment to be visible in your Jupyter notebooks.
5. Click: File > Hub Control Panel > Stop My Server > Start My Server
6. Reopen your notebook. Then click: Kernel > Change Kernel... and select the name of your custom conda environment from the drop-down list. |
|
4
|
Sat Feb 24 18:18:02 2024 |
Rutuja Gurav | Infrastructure | Computers | MATLAB on JupyterHub |
Followed the instructions in this installation guide - https://github.com/mathworks/jupyter-matlab-proxy - to install the jupyter-matlab-proxy in the jupyterhub python environment using command
sudo /opt/jupyterhub/bin/python3 -m pip install jupyter-matlab-proxy
But upon first execution I have to provide Mathworks accounts details.
The license on Chimay belongs to Prof. Jon. So, he'll have to insert those.
Right now MATLAB kernels for jupyter notebook as well as console show up (as shown in screenshot below) but code will not execute.
If I click on Open MATLAB tab, it will ask for Mathworks account login details.
The installation guide mentioned above says,
"When you execute MATLAB code in a notebook for the first time, enter your MATLAB license information in the dialog box that appears. See Licensing for details. The MATLAB session can take a few minutes to start."
Follow-up with Prof. Jon needed. |
| Attachment 1: richardsonlab_jupyter_matlab_proxy__user_rutuja.png
|
|
|
5
|
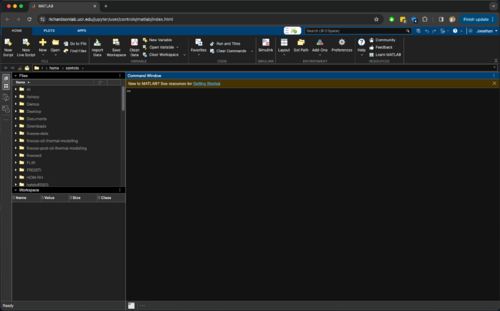
Sun Feb 25 08:27:00 2024 |
Jon | Infrastructure | Computers | MATLAB on JupyterHub |
| I activated the MATLAB license. All appears to be working now. The attached screenshot is what I now see upon clicking the Open MATLAB tab.
| Quote: |
Followed the instructions in this installation guide - https://github.com/mathworks/jupyter-matlab-proxy - to install the jupyter-matlab-proxy in the jupyterhub python environment using command
sudo /opt/jupyterhub/bin/python3 -m pip install jupyter-matlab-proxy
But upon first execution I have to provide Mathworks accounts details.
The license on Chimay belongs to Prof. Jon. So, he'll have to insert those.
Right now MATLAB kernels for jupyter notebook as well as console show up (as shown in screenshot below) but code will not execute.
If I click on Open MATLAB tab, it will ask for Mathworks account login details.
The installation guide mentioned above says,
"When you execute MATLAB code in a notebook for the first time, enter your MATLAB license information in the dialog box that appears. See Licensing for details. The MATLAB session can take a few minutes to start."
Follow-up with Prof. Jon needed. |
|
| Attachment 1: MATLAB_window.png
|
|
|
6
|
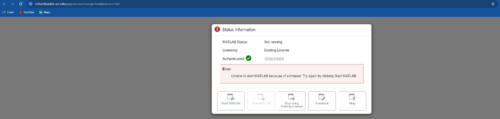
Sun Feb 25 14:06:48 2024 |
Rutuja Gurav | Infrastructure | Computers | MATLAB on JupyterHub |
I was incorrect about Prof. Jon needing to activate the MATLAB license for JupyterHub for all users to use the Jupyter MATLAB Proxy. Turns out the license on Chimay cannot be used by multiple users. But no worries, a user can click on Open MATLAB tab and then on the Online License Manager window login to your Mathworks account to use your UCR-provided license. If you instead click on Existing License tab and try to use the currently active license, your startup will eventually fail. See attached screenshots.
| Quote: |
| I activated the MATLAB license. All appears to be working now. The attached screenshot is what I now see upon clicking the Open MATLAB tab.
| Quote: |
Followed the instructions in this installation guide - https://github.com/mathworks/jupyter-matlab-proxy - to install the jupyter-matlab-proxy in the jupyterhub python environment using command
sudo /opt/jupyterhub/bin/python3 -m pip install jupyter-matlab-proxy
But upon first execution I have to provide Mathworks accounts details.
The license on Chimay belongs to Prof. Jon. So, he'll have to insert those.
Right now MATLAB kernels for jupyter notebook as well as console show up (as shown in screenshot below) but code will not execute.
If I click on Open MATLAB tab, it will ask for Mathworks account login details.
The installation guide mentioned above says,
"When you execute MATLAB code in a notebook for the first time, enter your MATLAB license information in the dialog box that appears. See Licensing for details. The MATLAB session can take a few minutes to start."
Follow-up with Prof. Jon needed. |
|
|
| Attachment 1: richardsonlab_jupyter_matlab_proxy_HowTo__user_rutuja__FAILED.png
|
|
| Attachment 2: richardsonlab_jupyter_matlab_proxy_HowTo__user_rutuja.png
|
|
|
9
|
Thu Feb 29 16:22:07 2024 |
Pooyan | Infrastructure | Computers | Malware incident |
Summary: There was a Cryptocurrency mining malware running on Chimay for ~2 days and using all of its CPU. Only one user was compromised, most probably by Brute-force ssh login. The malware, it's files, and the user was removed.
Log:
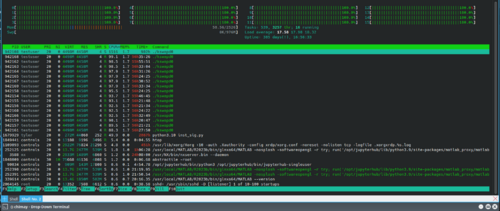
Two days ago I noticed a high CPU usage on Chimay. The proccess "kswapd0" run by user "testuser" was taking all of the ram. [htop screenshot attached]
I followed the answer here [https://askubuntu.com/a/530661] to make kswapd0 use swap only if there is no usable RAM left on the system.
also restarted the swap manually and changed the swappiness by sysctl, but the process was still there. Killing the process didn't work as it was back running in a few seconds.
I then came across this question [https://askubuntu.com/questions/1224927/cpu-100-with-kswapd0-process-although-no-swap-is-needed], showing that there is a coin miner malware called "Multios.Coinminer.Miner", hiding itself to look like the kswapd0 process.
Contamination by the malware is confirmed by looking at the directory that the code resides in the testuser directory. No other user seems to be infected. Logs show that this user was created three days ago and is only used for JupyterHub testing.
from /var/log/auth.log:
Feb 26 13:25:49 localhost sudo: rutuja : TTY=pts/167 ; PWD=/home/rutuja ; USER=root ; COMMAND=/usr/sbin/useradd --create-home testuser
...
Feb 26 18:22:17 localhost sshd[845020]: Failed password for testuser from 14.18.92.211 port 54280 ssh2
...
Feb 26 23:36:03 localhost sshd[934183]: Accepted password for testuser from 43.128.69.133 port 52690 ssh2
Apparently the user was compromised a few hours after its creation, and started its job a little bit later. Looking at the failed login logs show that there is a constant attack on Chimay of Brute-force ssh logins with dictionary usernames and passwords.
I killed al the processes by the testuser, deleted the malicious files and its home directory, and deleted the user itself. Thee seems to be no residues of it. No cronjob related to the malware is seen. CPU usage is back to normal.
Suggestions:
It seems that no other user accounts were compromised. But because there is these brute-force login attempts on Chimay, I have two suggestions:
Changing the passwords and having stronger ones, replacing ssh-keys (The user was running rsync, but probably they didn't care about the files that we have)
Disabling ssh login by password and asking everyone to use ssh-keys
|
| Attachment 1: Screenshot_20240229_114210.png
|
|